-
PDF
- Split View
-
Views
-
Cite
Cite
M. Laura Juárez, M. Gabriela Murúa, M. Gabriela García, Marta Ontivero, M. Teresa Vera, Juan C. Vilardi, Astrid T. Groot, Atilio P. Castagnaro, Gerardo Gastaminza, Eduardo Willink, Host Association of Spodoptera frugiperda (Lepidoptera: Noctuidae) Corn and Rice Strains in Argentina, Brazil, and Paraguay, Journal of Economic Entomology, Volume 105, Issue 2, 1 April 2012, Pages 573–582, https://doi.org/10.1603/EC11184
Close - Share Icon Share
Abstract
Spodoptera frugiperda (J.E. Smith) is composed of two genetically distinct strains, the so-called corn strain and the rice strain. Whether the two strains differ in their host use is unclear, because laboratory experiments have not been able to show consistent host performance or preference differences between them, and field studies showed high rates of hybridization, as well as some degree asymmetric host use. To determine the distribution of the two strains and their association with host plants, we collected fall armyworm larvae from different crops (corn, rice, alfalfa, and sorghum) and grasses in 15 different localities over 4 yr in Argentina, Brazil, and Paraguay. The strain identity was analyzed using two polymorphisms in the mitochondrial cytochrome oxidase subunit I gene. We identified the corn and rice haplotypes and three types of populations were characterized based on the frequencies of the individuals that belonged to any of these haplotypes: in 44% of populations the corn haplotype predominated, in 44% of populations the rice haplotype was the most frequent, and 11% of populations showed both haplotypes at similar proportions. In total, eight populations (47%) showed the expected pattern, two populations (12%) were polymorphic within the same field, and seven populations (41%) showed the inverse pattern. Taken together, there was no consistent pattern of host association between the two sympatric genotypes and their respective host plants. This investigation supports the need for additional studies to determine which other forces keep the genotypes separate, and what is the degree of genetic differentiation between these populations.
Polyphagous insects may undergo population structuring processes throughout restricted mobility and selection for specialization to particular host plants, leading to what is called a host race. As such, host races or strains are intermediate stages in the continuum between population polymorphism and fully distinct species, representing an incipient stage in the process of sympatric speciation (Berlocher and Feder 2002, Drés and Mallet 2002). The existence of host strains, and the factors that contribute to their maintenance in nature, can have an important impact on pest control; particularly to delineate effective strategies for Bt crop resistance management.
The fall armyworm, Spodoptera frugiperda (J.E. Smith), is a polyphagous species that infest ≈186 host plants throughout the Western Hemisphere (Casmuz et al. 2010) including many agricultural crops (Sparks 1979, Andrews 1980, Betancourt and Scatoni 1996). It is an endemic pest of the American continent distributed from southeastern Canada to Chile and Argentina, including the Caribbean Islands, where it is able to survive all year around in tropical areas of Central and South America, Mexico, and subtropical environments of southern Florida and Texas (Mitchell 1979, Sparks 1979, Pair et al. 1986, Raulston et al. 1986). Being a nondiapausing species, in temperate and cold zones it behaves as a seasonal pest (Pair et al. 1991).
In Northeast Argentina, S. frugiperda is the most important pest of corn causing yield losses, fluctuating from 17 to 72% (Perdiguero et al. 1967; Willink et al. 1993a,b) and this region could be considered the southern limit of permanent distribution of the pest. Murúa and Virla (2004a) determined the presence of individuals during the winter months, suggesting that permanent populations should reach the province of Tucuman in Argentina (between 26° and 28° S). The same authors demonstrated the presence of larvae of S. frugiperda in different grasses during the spring in the absence of corn, indicating that populations that affect this crop may not depend solely on migration of individuals from other regions (Murúa and Virla 2004b).
Two host strains have been described for S. frugiperda: the so-called corn strain and the rice strain (Pashley et al. 1985, Pashley 1986).The corn strain has been reported to infest corn, Zea mays L., sorghum, Sorghum bicolor (L.) Moench subsp. Bicolor, and cotton, Gossypium hirsutum L., while the rice strain has been found mostly in rice, Oriza sativa L., sugar cane, Saccharum officinarum L., and grasses such as Johnson grass, Sorghum halepense (L.) Persoon, and Bermuda grass, Cynodon dactylon (L.).
The occurrence of these two morphologically indistinguishable strains has been confirmed in several regions in North and South America using molecular markers (Busato et al. 2004; Prowell Pashley et al. 2004; Martinelli et al. 2006; Nagoshi et al. 2006a, 2007a; Clark et al. 2007; Machado et al. 2008; Murúa et al. 2008; Vélez-Arango et al. 2008; Virla et al. 2008). The strains have been reported to exhibit mechanisms of reproductive isolation, developmental and physiological variations, and genetic polymorphisms (Pashley 1986, 1988; Pashley and Martin 1987; Whitford et al. 1988; Quisenberry 1991; Pashley et al. 1995; Adamczyk et al. 1997; McMichael and Prowell 1999; Nagoshi and Meagher, 2004; Groot et al. 2010). However, the results from different studies, which in some cases included hybrid progeny, were not always consistent, with no physiological or behavioral studies under laboratory conditions being able to distinguish between the two strains.
In terms of reproductive isolation, different premating mechanisms have been found between the two strains, including oviposition preference (Whitford et al. 1988), the temporal partitioning of nocturnal mating activities (Pashley et al. 1992, Schöfl et al. 2009) and sex pheromone differences (Groot et al. 2008, Lima and McNeil 2009). Postmating isolation studies are more conflicting. Pashley and Martin (1987) reported unidirectional reproductive isolation with no successful matings occurring between females of the corn strain and males of the rice strain, while the reciprocal cross produced fertile F1 progeny. Similar results were found with Mexican populations (López-Edwards et al. 1999) and Argentinean populations (Murúa et al. 2008). In contrast, Whitford et al. (1988), Quisenberry (1991), Nagoshi and Meagher (2004), and Groot et al. (2010) were able to obtain fertile progeny from interstrain mating in both directions. However, in backcrosses, the hybrid females from crosses between females of the rice strain and males of the corn strain were less fertile; either mating parental did not occur, or females produced fewer fertile egg masses and the interhybrid matings reproduced at a lower rate (Pashley and Martin 1987, Whitford et al. 1988, Groot et al. 2010). Nagoshi and Meagher (2004) determined that laboratory culturing and conditions may confound strain specific mate selection, and Groot et al. (2010) suggested that the geographic origin of the strain may affect hybridization success in any of the two reciprocal cross directions.
In terms of larval performance, the rice strain was found to develop better on rice plants than on corn plants, while the corn strain performed equally well on both plants (Pencoe and Martin 1981, Pashley 1988, Whitford 1988, Pashley et al. 1995). Conversely, in nature the corn strain seems to be more restricted to corn habitats and is rarely found in rice habitats, while the rice strain can be found on corn as well (Pashley et al. 1995). In addition, Meagher et al. (2004) and Groot et al. (2010) found that rice strain larvae were heavier and developed faster on corn and sorghum-sudangrass than corn strain larvae. These results showed that the larval performance does not seem to translate into differential fecundity and fertility (Pashley et al. 1995). Moreover, field sampling from several regions showed high rates of hybridization as well as some degree of asymmetric host use according to evidence from several molecular markers (Prowell Pashley et al. 2004). The hybrid genotypes mostly consisted of rice females and corn males (Nagoshi and Meagher 2003b, 2004; Prowell Pashley et al. 2004). Prowell Pashley et al. (2004) showed that hybrids mostly occurred in corn habitats. However, Nagoshi et al. (2006b) found that the two habitats differed in the proportions of the parental genotypes. Rice habitats contained mostly individuals with rice mitochondrial haplotype (mtR) and a nuclear FR tandem-repeat sequence, present in rice and absent in corn (FR+) and mtC FR0 in corn habitats, confirming the preferences of strains to their habitats. In contrast, mtR FR0 (rice female and corn male) hybrids showed no habitat bias. The authors concluded that this lack of bias reflects that the host preference is compromised by the mixing of genomes.
The two strains show allozyme polymorphisms (Pashley 1986), nuclear genome polymorphisms (Lu et al. 1992, 1994; McMichael and Prowell 1999; Prowell Pashley et al. 2004; Nagoshi and Meagher 2003a,b; Busato et al. 2004; Martinelli et al. 2006, 2007; Clark et al. 2007; Machado et al. 2008) and mitochondrial DNA polymorphisms (Lu and Adang 1996; Levy et al. 2002; Meagher and Gallo Meagher 2003; Prowell Pashley et al. 2004; Nagoshi et al. 2006a, 2007a,b; Vélez Arango et al. 2008; Virla et al. 2008). Among these molecular markers, the most consistent and convenient for distinguishing between the strains are the mitochondrial haplotypes that were shown to differentiate the two strains in their sequence of DNA in the cytochrome oxidase I (COI) gene (Lu and Adang 1996, Levy et al. 2002, Nagoshi et al. 2006a). The MspI restriction site is present in the corn strain but not in the rice strain, while the SacI restriction site is present in the rice strain and not in the corn strain. This double digestion was developed as an effective tool for the identification of the two strains, and showed a strong correlation with the preferred host plant (Lu and Adang 1996, Levy et al. 2002, Meagher and Gallo-Meagher 2003, Nagoshi et al. 2006a).
Whether and to what extent the two host strains are associated to specific host plants has a significant implication both from a theoretical and applied perspective. If S. frugiperda strains are not associated to specific host plants, then the mechanisms that maintain genetic structuring in nature are not host dependent, providing an interesting study model to elucidate evolutionary pathways. From an applied perspective, the presence of a strict host-strain association would restrict the alternative hosts that can be used as refuges to prevent or delay emergence of resistant individuals if Bt crops are widely grown. In areas with genetically modified Bt corn, where it is necessary to maintain susceptible populations (Georghious and Taylor 1977, Tabashnik and Croft 1982, Caprio 2001, Bourguet et al. 2003) the current strategy is the use of refuges, that is, areas planted with conventional corn in the same site as Bt corn, to act as a source of susceptible individuals. However, other crops or even grasses or pastures have been proposed as alternatives to nontransgenic corn to act as refuge areas (Gould 1998, Green et al. 2003, Murúa et al. 2009).
The objective of this study was to determine whether the two S. frugiperda strains described in other regions are present in different crops in three countries of South America, with an emphasis on Argentina, and to verify or dispel the association of the strains with their putative host plants. For the identification of the two strains we used the mitochondrial haplotypes, where the two strains differ in their sequence of DNA in the COI. Because we only used these mitochondrial markers to identify all individuals in this study, we will refer to the individuals as corn mitochondrial haplotypes and rice mitochondrial haplotypes.
Materials and Methods
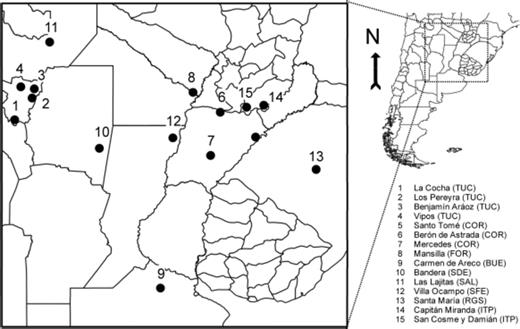
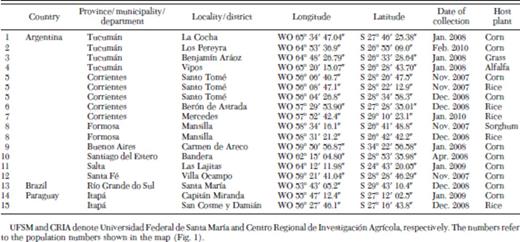
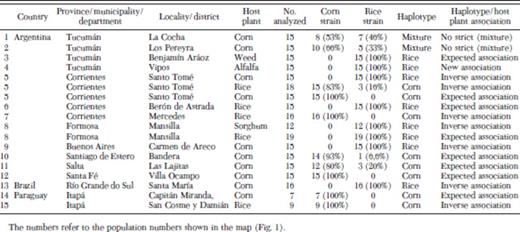
Fall armyworm larvae were collected from different crops and localities in commercial fields from seven Argentinean provinces (Table 1; Fig. 1): Tucumán (TUC), Corrientes (COR), Buenos Aires (BUE), Salta (SAL), Santiago del Estero (SDE), Santa Fé (SFE); and Formosa (FOR); one Brazilian municipality, Santa María in the state of Río Grande do Sul (RGS), and two Paraguayan districts, Capitán Miranda and San Cosme y Damián in the department of Itapúa (ITP). Collections took place in November 2007, December 2008, December 2009, and January 2010.
Localities and host plants sampled in different years from Argentina, Brazil, and Paraguay for S. frugiperda haplotypes identification
 |
 |
Localities and host plants sampled in different years from Argentina, Brazil, and Paraguay for S. frugiperda haplotypes identification
 |
 |
Localities and provinces of the S. frugiperda sampling sites in Argentina (TUC, Tucumán; SAL, Salta; SDE, Santiago del Estero; SFE, Santa Fé; FOR, Formosa; BUE, Buenos Aires; COR, Corrientes), Brazil (RGS, Rio Grande do Sul), and Paraguay (ITP, Itapúa). The numbers refer to the population numbers shown in Table 1 and 2.
At each sample site, larvae of all instars (a minimum of 250) were collected from commercial fields. The crops sampled were corn, rice, sorghum, alfalfa (Medicago sativa L.) Bermuda grass, and Guinea grass (Megathyrsus maximum (Jacq.)). The last two host plants will be referred to as grasses in this study. The collected larvae were placed individually in glass tubes (12 cm high and 1.5 cm diameter) with leaves of the host plant. Larvae were taken to the laboratory and placed in chambers at 27 ± 2°C, 70–75% RH and a 14:10 L:D photoperiod until pupation and adult emergence. Late instar larvae and adults were examined to confirm that all individuals were fall armyworm based on diagnostic taxonomic characters. Voucher specimens were deposited in the collection of Sección Zoología Agrícola, Estación Experimental Agroindustrial Obispo Colombres, Tucumán, Argentina.
Populations from each sampled crop in each province were maintained separately, and 200 adults were used from each population to establish laboratory colonies. In separate mating cages (30 cm high and 10 cm diameter cylindrical polyethylene-terephthalate cages with nylon mesh cloth) 4–5 < 24 h adults ♀ and 4–5 adults ♂ were introduced. We had in total ≈20 mating cages per population. The cages contained pieces of paper that allowed females to rest and to lay eggs. Food was provided via a cotton plug saturated with a mixture of honey and water (1:1 vol:vol), which was renewed every day. Cages were checked daily for oviposition and adult mortality. To minimize loss of genetic variability, once females started to lay egg masses, ∼15 egg masses from each cage were collected and deposited in glass tubes (12 cm high and 1.5 cm diameter). Once emerged, 15 neonate larvae from each of the different egg masses were placed individually in glass tubes with artificial diet (Osores et al. 1982), which was renewed every 2–3 d. As larvae pupated, they were placed in cylindrical cages until adult emergence. On average, 200 adults were used again to initiate a new generation. After establishing a colony from each population and crop, larvae from the second generation were stored at −20°C until DNA extraction and used for the following analysis.
Total DNA was extracted from fall armyworm larvae (≈100 mg) that were ground in liquid nitrogen. The remainder of the extraction procedure was performed using a CTAB method except for the addition of proteinase K and the final resuspension of the DNA was in 70 μl of bi-distilled water. Quality and concentration of DNA were determined by comparison of DNA bands observed in agarose gels (0.1% agarose gel stained with 0.5 μg/ml ethidium bromide) with standard DNA bands of known concentrations (100–1000 ng). Buffer and running conditions were performed according to Sambrook et al. (1989).
We used two restriction site length polymorphisms, the MspI and SacI polymorphism in COI, to characterize the two mitochondrial haplotypes of the fall armyworm. Primers were synthesized by Sigma-Aldrich (Buenos Aires, Argentina) and included the pairs JM 76 (5′-GAGCTGAATTAGG(G/A)ACTCCAGG-3′) and JM 77 (5′-ATCACCTCC(A/T)CCTGCAGGATC-3′) (Levy et al. 2002).
Polymerase chain reaction (PCR) amplification of the mitochondrial COI gene was performed in a 25 μl reaction mix containing 1X reaction buffer Go Taq, 2 mM MgCl2, 0.01 mM dNTPs, 0.001 μM primers, 1 U of TaqDNA polymerase (Promega, Madison, WI) and 80 ng DNA total. The thermocycling program was an initial incubation at 100°C, 1 min followed by 30 cycles: denaturation, 1 min at 94°C; annealing, 1 min at 58°C; extension, 7 min at 72°C with a final segment of 7 min at 72°C. The reaction was carried out in a MJ Research Thermal Cycler (PTC-100; MJ Research, Inc., Watertown, MA). Negative controls, without DNA, were included in each experiment. At the end of the PCR, amplification products were separated by electrophoresis in 1.5% agarose gels and these were stained with ethidium bromide. Molecular weights were estimated using 100 bp PCR markers from Promega.
The PCR-amplified products were purified using the protocol of Perera et al. (2009). After purification, 5 μl of each sample was digested with either MspI and SacI restriction endonuclease, in separate reactions. Samples were incubated at 37°C for 3 h and digestion products were separated by electrophoresis in 2% agarose gels. Negative controls, without enzymes, were included in each experiment.
We considered the populations to belong to one of the two haplotypes if the frequency of the corresponding haplotype was above 80% (see Table 2). When haplotype frequencies were 0.8 < P < 0.2, we considered the population as a mixture of haplotypes.
mtDNA analysis showing the presence of the haplotypes types based on frequency of occurrence in S. frugiperda populations collected from Argentina, Brazil, and Paraguay
 |
 |
mtDNA analysis showing the presence of the haplotypes types based on frequency of occurrence in S. frugiperda populations collected from Argentina, Brazil, and Paraguay
 |
 |
Results
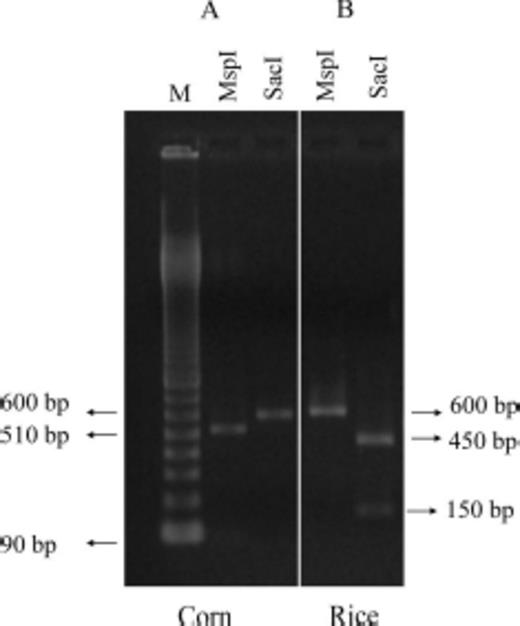
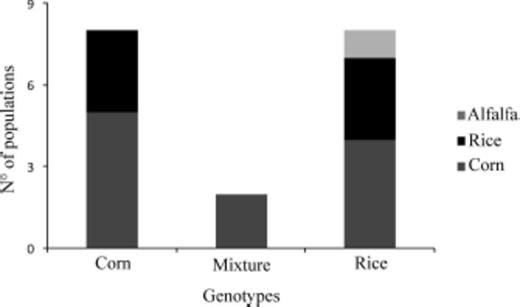
The fragments produced by PCR amplification with JM 76 and JM 77 primers was 600 bp, which were separately digested with MspI and SacI restriction enzymes. The PCR-amplified products from the corn mitochondrial haplotypes were cut once by MspI producing two fragments of 510 and 90 bp in length, but not by SacI. DNA products from the rice mitochondrial haplotypes showed a reciprocal pattern: they were cut by SacI producing two fragments of 450 and 150 bp, but not by MspI (Fig. 2). Thus, we identified two distinct genotypes and three distinct types of populations were characterized based on the frequencies of the individuals that belonged to these haplotypes: in 44% of populations the corn haplotype predominated, in 44% of populations the rice haplotype was the most frequent, and 11% of populations showed both haplotypes at relatively similar proportions (Table 2; Fig. 3).
Agarose gel displaying PCR- amplified fragments of COI mitochondrial region of S. frugiperda larvae produced by the JM 76/JM77 primers pair and digested with MspI and SacI enzymes. (A) Samples belonging to corn strain. (B) Samples belonging rice strain. The uncut fragment is 600 bp and M denote molecular marker.
Genotypes of S. frugiperda populations collected from different hosts. Corn genotype includes five populations collected from corn (gray bar) and three populations collected from rice (black bar). Mixture of genotypes includes two populations collected from corn (gray bar). Rice genotype includes two populations collected from rice and one population collected from grass (black bar), three populations collected from corn and one population collected from sorghum (gray bar), and one population collected from alfalfa (light gray bar).
The results from the 18 populations sampled in nine different provinces or districts are presented in Table 2. In Argentina we found both mitochondrial haplotypes, in Paraguay we found only the corn mitochondrial haplotype and in Brazil we found only the rice mitochondrial haplotype. In terms of haplotype–host association, three situations were found, that is, expected association, mixture of haplotypes, and inverse association.
Expected associations (corn haplotypes in corn habitats and rice haplotypes in rice habitats) were found in 8/17 of the populations (Table 2; Fig. 3). The population collected from grasses in Tucumán in 2008, from corn and rice in Corrientes in 2008, from rice in Formosa in 2008, from corn in Santa Fé, Santiago del Estero, and Salta (Argentina) in 2007, 2008, and 2009, respectively and from corn in Capitan Miranda (Paraguay) in 2009. In Tucumán, individuals collected from grasses belonged to the rice haplotype and this was expected given the fact that in this region rice is not cultivated, grasses represent the only host available for a putative rice haplotype. Thus, expected associations were found in different regions and in different years.
No clear associations were found in 2/17 populations from Argentina (Table 2; Fig. 3). In Tucumán, on two occasions during the 2 yr of sampling, larvae collected from corn showed a mixture of genotypes. As mentioned before, in this province, rice plants are not cultivated, hence pastures and grasses could serve as a host plant for the rice strain (Pashley 1986, Lu and Adang 1996, McMichael and Prowell 1999, Levy et al. 2002, Prowell Pashley et al. 2004).
Nonexpected associations (corn haplotypes in rice habitats and rice haplotypes in corn habitats) were found in a total of 7/17 populations, collected in Argentina, Brazil, and Paraguay (Table 2; Fig. 3). In the provinces of Buenos Aires, Corrientes (in Santo Tomé in 2007) and Formosa (in Mansilla in 2007), all the individuals belonged to the rice haplotype, although they were sampled in corn and sorghum fields, respectively. In addition, in Santa Maria (Brazil) the 16 individuals analyzed from corn belonged to the rice haplotype. Interestingly, in this area rice is cultivated extensively, but no larvae were found in four different rice fields (Palmas, Séptimo distrito, Varzea do Agudo, and São Sepé). In 2007 and 2010 in Corrientes (in the localities of Santo Tomé and Mercedes, respectively), all individuals collected from rice were typed as belonging to the corn haplotype. The same was observed in San Cosme y Damián.
With respect to the population collected from alfalfa in the province of Tucumán, all individuals corresponded to the rice haplotype. To our knowledge, this is the first time S. frugiperda larvae from alfalfa have been identified to strain, so that we did not have any previous expectation for this crop.
Discussion
The current study aimed at determining the existence of the two S. frugiperda strains previously characterized (i.e., corn and rice strains) and assess a correlation between their occurrence and the host plants from which they were collected. We consistently found the two haplotypes described as diagnostic for rice and corn strains and based on the frequency of occurrence within the analyzed South American populations, three types of populations were found: two that bear one haplotype fixed or at high frequency (higher than 0.8) and the third one polymorphic with both haplotypes at intermediate frequencies (0.2 < P < 0.8). In total, only eight populations (47%) showed the expected pattern, two populations (12%) were polymorphic at mtDNA level within the same field, and the remaining seven populations (41%) showed the inverse pattern. Taken together, there was no consistent pattern of host association between the two sympatric distinct genotypes sampled in the area and their respective host plants.
Although we assume that our laboratory populations represent the mitochondrial haplotypes of wild populations, there is a possible effect of homogenization as a result of our laboratory rearing (Berlocher and Friedman 1981; Simões et al. 2008, 2010). To avoid this, we collected a large number of individuals from wild populations that were used to establish the colonies, and the material for DNA extraction was obtained from the very first generations in culture. Although the possible bias associated with sampling error is the underestimation of haplotypic diversity, the two haplotypes have been found to occur in almost all regions where S. frugiperda populations were characterized. Therefore, we have no evidence of homogenization effect that could have produced a predominance of one mitochondrial haplotype at a regional level. In other words, there is a low probability that the same allele was fixed twice in different years, for example in Corrientes in 2007 and 2010. In addition, two populations collected from corn fields in Tucumán in two different years showed no tendency toward dominance of one haplotype, but clearly a mixture of the two haplotypes.
Despite the fact that we found no clear association between the two haplotypes and the host plants from which they were collected, our findings are not that different from previous studies. In general, ≈ 80% of collected individuals from corn genotype habitats were found to belong to the corn genotype, while the other 20% belonged to rice genotype (Lu and Adang 1996, McMichael and Prowell 1999, Prowell Pashley et al. 2004, Nagoshi et al. 2007a, Machado et al. 2008). We found a similar “infidelity” of the rice mitochondrial haplotype, being dominant in four populations collected from corn habitats. The rice genotype apparently has a wider host plant distribution and can be found in more habitats than only rice (e.g., in alfalfa). In the case of the corn haplotype, it was unusual to find three predominantly corn haplotype populations from Corrientes and Paraguay in rice habitats, because generally ≈ 85–90% of collected larvae in rice from other regions has previously been identified as rice genotype (Lu and Adang 1996; Nagoshi and Meagher, 2003b, 2004; Machado et al. 2008; Vélez Arango et al. 2008). Our finding of 100% corn individuals in rice habitats has not been reported so far and this could be because rice fields were not extensively sampled as corn fields in previous studies. Hence, our results provide new data about the fall armyworm genotype distributions on this crop.
The fact that we found both mitochondrial haplotypes in both habitats reveals biologically interesting aspects of what may be occurring in these South American populations: 1) The two haplotypes may be in an early process of divergence, constituting a polymorphic condition with the main differences at haplotype frequencies levels, where the variants have not reached fixation in the populations. This may result in the fact that the molecular pattern observed in other regions with this technique cannot be extrapolated to the entire distribution of the species. 2) The mitochondrial haplotypes may exhibit plasticity in their host usage, probably because of environmental constraints. 3) The mitochondrial haplotypes may have diverged because of a host shift, but they are still adapted to both host plants and other mechanisms maintain the integrity of different genotypes irrespective of which host is used by the larvae. 4) Some populations are hybrids in which the linkage between the COI marker and the strain specificity has been broken or this mitochondrial marker, unlike previous reports, is insufficient to detect the association with the host.
Murúa et al. (2008) showed unidirectional incompatibility between populations from the north of Argentina with a population from Buenos Aires. Females from North Argentinian populations crossed with males from Buenos Aires showed a lower number of spermatophores as compared with homotypic crosses. There was also a tendency to reduced egg viability in heterotypic crosses where spermatophores were found. These observations led the authors to postulate that the North Argentinian population was the corn strain and the Buenos Aires population belonged to the rice strain. However, no strain typing on these populations was conducted. As a follow up, Pereyra (2009) conducted reproductive compatibility studies between samples from grasses, alfalfa, and corn from Tucumán, but did not observe any mating incompatibility among them. The difference between these two studies is that in the first case populations are geographically separated (≈1,200 km apart), while the latter populations are relatively close (≈100 km apart). Thus, it is possible to assume in the first case the occurrence of genetic differentiation attributable to isolation by distance. Northeastern Argentina (e.g., Corrientes and Formosa) is considered the country's rice area, where large areas of this crop are planted every year, in addition to corn and sorghum, among others. During the years of sampling, we found individuals in all these crops, however, the dynamics of the pest populations tends to fluctuate in different region. For example, in a province next to Corrientes that we monitored, Entre Rios, we were unable to find larvae on rice, only on corn. It was not until later months and possibly related to the lack of availability of corn that we found individuals on rice. The same possibly occurs in the province of Buenos Aires, where corn sowing is delayed compared with other areas and there are no rice crops. For this reason and because the weather conditions are not favorable, this area probably consist of nonpermanent populations being colonized every year by individuals from other regions (northeastern of Argentina, from Brazil or Uruguay) (Murúa et al. 2008). Therefore, one would expect that individuals migrating to this area would belong to both strains. This may be another explanation for the occurrence of both mitochondrial haplotypes, that is, maybe they differ in their migration patterns or the preference of haplotype to different host plants may be influenced by environmental and seasonal factors exhibiting differences in the availability of alternative host plants as was determined by other studies (Prowell Pashley et al. 2004; Nagoshi et al. 2006a, 2007a,b; Machado et al. 2008; Vélez Arango et al. 2008). Interestingly, however, the Buenos Aires population from corn fields analyzed by Nagoshi et al. (2012) may be hybrids, because it was typified as rice haplotype with the mitochondrial COI marker, but as corn haplotype by its nuclear TPI gene sequence.
Given that we used a mitochondrial marker, which has maternal inheritance, we cannot determine whether we were dealing with hybrid populations. In another study, the combined use of mitochondrial and nuclear markers revealed a discordance frequency of 16% in fall armyworm individuals for at least one marker (Prowell Pashley et al. 2004). The hybrids mostly resulted from crosses of rice females (as shown by the mitochondrial marker) with corn males (shown by the nuclear AFLP markers and allozymes). The same was found by Nagoshi and Meagher (2003b, 2004), using mitochondrial haplotypes and the nuclear tandem-repeat sequence (FR). Their results indicate hybridization between rice strain females and corn strain males as well. Such hybridizations may explain why we found mostly rice strain individuals in corn strain habitats: the mitochondrial marker would diagnose these hybrids as rice strain. However, such a high incidence of hybrids would quickly result in a homogenization of the two strains, which makes it unlikely that all are hybrids. On the other hand, Prowell Pashley et al. (2004) also postulated that discordant individuals could be the results of ancestral polymorphism where there was no fixation of alleles, so that the same variant may exist in both genotypes. Although this hypothesis was rejected on the basis of biological and genetic evidence observed in North American and Caribbean populations (Pashley and Martin 1987; Pashley et al. 1995; Nagoshi and Meagher 2003b, 2006b), we cannot discard this possibility for the South American populations that we describe here.
Now that we found that both mitochondrial haplotypes are present in these regions, and that they are not restricted to specific habitats, it is important to clarify whether another evolutionary process may be occurring in South American populations, or if the presence of different agro-ecosystems and different management systems may contribute to differences in the genetic structure of the populations. To solve this question, it is necessary to assess the genetic structure between the two strains from similar and different host plants with other molecular markers and corroborate these associations for efficient pest management strategies. The lack of association between strains and host plant with the high polyphagy, and the ability of the pest to spread and move from different Bt-crops to surrounding crops and grasses, and vice versa, could imply that refuge areas may not be necessary in certain conditions. These crops (especially corn and rice) would be a source of individuals of both strains creating the conditions to prevent the development of resistance in areas where Bt crops are found in large areas. However, this is a hypothesis that needs testing. Therefore, further biological and population genetic studies are still necessary to elucidate the gene flow between populations in different habitats with additional molecular techniques that can detect pure and hybrid strains, to assess unequivocally whether the two strains use different hosts with no specific association and thus have a better understanding and application to pest management.
In conclusion, our results from mtDNA provide new information on the distribution of the fall armyworm haplotypes in South America. In general, we found no consistent pattern of host association for either of the two distinct haplotypes across years and regions, showing that the host plant–haplotype association is not strict, at least in Argentina. The lack of clear host-haplotype associations revealed in our study indicates that the different genotypes probably maintain their integrity through other mechanisms, independent of the host in which the larva fed, such as differential timing of mating at night, differences in sex pheromones of females from both strains (Pashley et al. 1992, Groot et al. 2008, Lima and McNeil 2009, Schöfl et al. 2009) and postmating incompatibilities (Pashley and Martin 1987, Whitford et al. 1988, López-Edwards et al. 1999, Murúa et al. 2008, Groot et al. 2010). Further research is necessary to determine which of these factors if any maintain the integrity of the two strains, and whether there is some difference in the migratory behavior of the pure strains and their hybrids.
We are grateful to Jerson Guedes (Universidad de Santa María, Río Grande do Sul, Brazil) and Nancy Espinoza (Centro Regional de Investigaciones Agrarias, Itapúa, Paraguay) for their collaborations in the sampling. We acknowledge Juan Rull (Instituto de Ecología, Xalapa, Mexico) for his comments and critical review of an earlier version of the manuscript. We are also very grateful for all suggestions and comments provided by the reviewers throughout the revision process. Finally, we thank Pablo Scandaliaris and Silvio Salmoiraghi (EEAOC) for his assistance in the preparation of the figures and Fernando Navarro (Facultad de Ciencias Naturales e IML, Universidad de Tucumán) for the taxonomic identification of S. frugiperda larvae.
References Cited